RESEARCH
1. ONCOPROTEOMICS
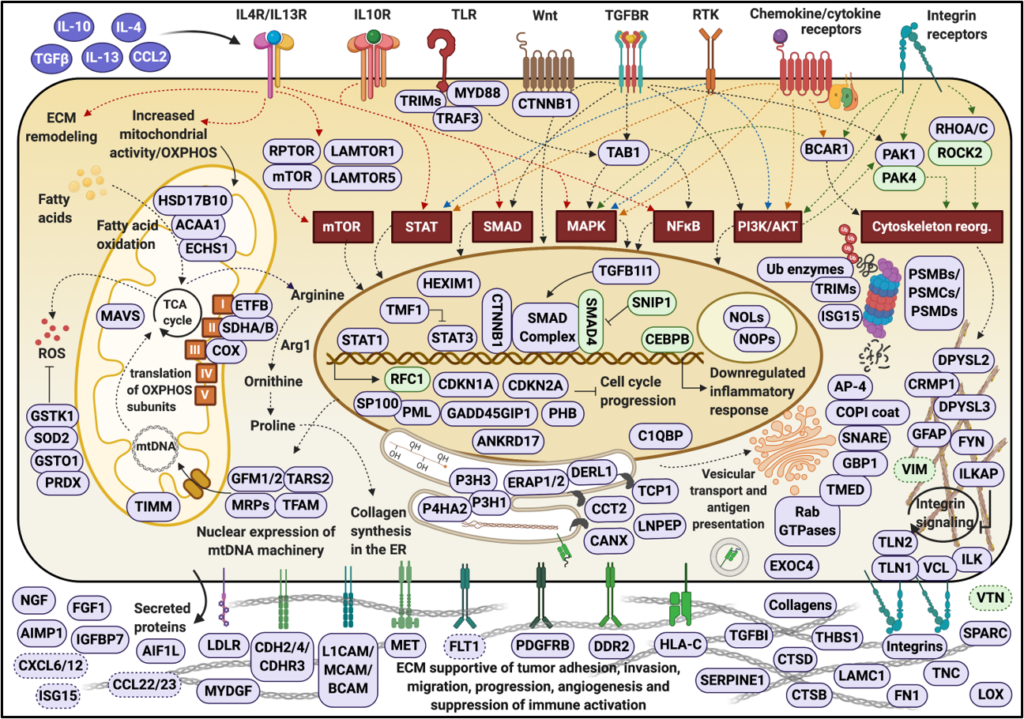
Cancer is a disease of the cell cycle that results in uncontrolled proliferation of cells. Most cancer phenotypes exhibit some, if not all, of eight major characteristics: ability to evade apoptosis, proliferate in the absence of growth factors, ignore inhibitory factors, recruit their own blood supply, avoid immune destruction, display deregulated cell energetics, become immortal and metastasize. In our laboratory, we explore the molecular mechanisms of cancer cell cycle regulation and metastatic potential by using holistic, mass spectrometry-based systems biology approaches. We develop proteomic technologies for investigating the pathways that enable cancer cells to bypass tightly regulated molecular checkpoints, proliferate in an unrestrained manner, metastasize and hijack normal biological function. We perform comprehensive proteomic profiling of cancerous cells, and develop stable isotope labeling and label-free quantitative approaches for differential protein expression profiling of complex cellular extracts.
- Ahuja, S.; Lazar, I.M., “Proteomic Insights into Metastatic Breast Cancer Response to Brain Cell-Secreted Factors,” Sci. Rep. 2024, 14, Article 19351. https://doi.org/10.1038/s41598-024-70386-7
- Ahuja, S.; Lazar, I.M., “Systems-Level Proteomics Evaluation of Microglia Response to Tumor-Supportive Anti-inflammatory Cytokines,” Front. Immunol., 2021, 12, Article 646043. https://doi.org/10.3389/fimmu.2021.646043
- Lazar, I.M.; Hoeschele, I.; De Morais, J.A.; Tenga, M.J., “Cell Cycle Model System for Advancing Cancer Biomarker Research,” Sci. Rep. 2017, 7, 17989 (12 p.). https://doi.org/10.1038/s41598-017-17845-6
- Tenga, M.J.; Lazar, I.M., Proteomic Snapshot of Breast Cancer Cell Cycle: G1/S Transition Point,” Proteomics 2013, 13(1), 48-60. https://doi.org/10.1002/pmic.201200188
- Armenta, J.M.; Hoeschele, I.; Lazar, I.M., “Differential Protein Expression Analysis Using Stable Isotope Labeling and PQD Linear Ion Trap MS Technology,” Am. Soc. Mass Spectrom. 2009, 20, 1287-1302. https://doi.org/10.1016/j.jasms.2009.02.029
2. CANCER DRUG TARGETS, BIOMARKERS AND METASTASIS
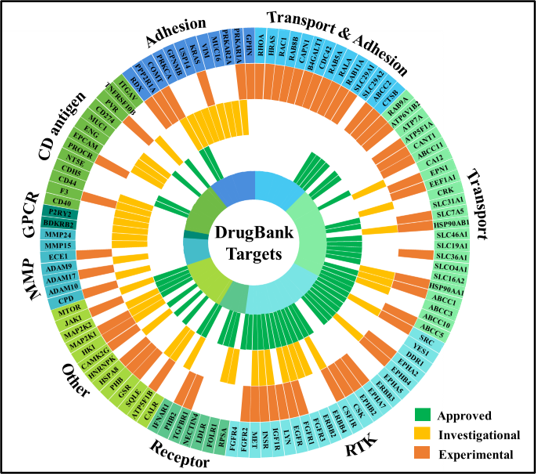
By providing novel insights into the functional protein networks that drive cancer cells into division, our data point to a broad range of potential therapeutic targets that concurrently affect the cell cycle signaling and transcriptional/translational machinery. The identified proteins can be used alone, or in combination, as target molecules for developing novel anti-cancer drugs and therapies, or as disease signatures for advancing cancer detection and diagnosis. Our results indicate that many molecular markers detected in breast cancer cells are not specific, but common to multiple cancers including brain and various neurological disorders. By capitalizing on these data, we further aim at identifying the signaling pathways that increase metastatic potential, and the protein networks with prognostic value for brain metastasis.
- Karcini, A.; Mercier, N.R.; Lazar, I.M., “Proteomic Assessment of SKBR3/HER2+ Breast Cancer Cellular Response to Lapatinib and Investigational Ipatasertib Kinase Inhibitors,” Front. Pharmacol. 2024, 15. https://doi:10.3389/fphar.2024.1413818
- Karcini, A.; Lazar, I.M., “The SKBR3 Cell-Membrane Proteome Reveals Telltales of Aberrant Cancer Cell Proliferation and Targets for Precision Medicine Applications,” Rep. 2022, 12, Article 10847. https://doi.org/10.1038/s41598-022-14418-0
- Lazar, I.M., Karcini, A., Haueis, J.R.S, “Mapping the Cell-Membrane Proteome of the SKBR3/HER2+ Cell Line to the Cancer Hallmarks,” PLOS ONE 2022, 17(8), Article e0272384. https://doi.org/10.1371/journal.pone.0272384
- Lazar, I.M., Kontoyianni, M., Lazar, A.C., Eds., “Proteomics for Drug Discovery,” book in the series of Methods in Molecular Biology, Springer, 2017. (ISBN 978-1-4939-7201-2) https://link.springer.com/book/10.1007/978-1-4939-7201-2
- Tenga, M.J.; Lazar, I.M., “Proteomic Study Reveals a Functional Network of Cancer Markers in the G1-Stage of the Breast Cancer Cell Cycle,” BMC Cancer 2014, 14, 710. https://doi.org/10.1186/1471-2407-14-710
3. MICROFLUIDIC DEVICES FOR BIOMARKER DISCOVERY AND BIOLOGICAL SIGNALING
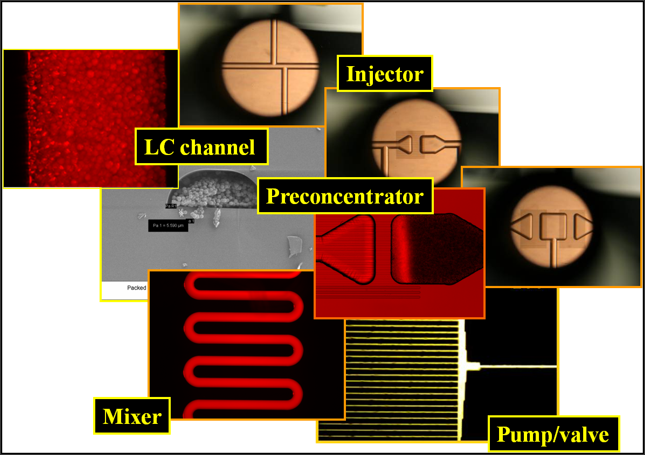
Our laboratory specializes in the development of microfluidic devices as a front-end to mass spectrometry detection. We are interested in the advancement of microfabricated platforms that encompass pressure and electrically driven fluid propulsion elements for pumping/valving and mixing, microreactors for protein enrichment and enzymatic digestion, and chromatographic separation systems for nano-flow capillary separations of complex mixtures. Our unique interest is the development of microchip-MS interfaces that enable electrospray ionization (ESI) and matrix assisted laser desorption ionization (MALDI) detection from a chip, and the integration of these interfaces into fully integrated, stand-alone microfluidic platforms for high-throughput proteomic investigations. We further build on the power of our proteomic data to develop microfluidic architectures for the detection of disease biomarkers and for monitoring fast signaling processes in cells. We integrate nano-particle sensing/reporter elements for studying cell behavior and understanding tumor development.
- Lazar, I.M.; Deng, J.; Stremler, M.A.; Ahuja, S., “Microfluidic Reactors for Advancing the MS Analysis of Fast Biological Responses,” Microsystems & Nanoengineering 2019, 5, 7 (16 pages). http://doi.org/10.1038/s41378-019-0048-3
- Deng, J.; Ikenishi, F.; Smith, N.; Lazar I.M., “Streamlined Microfluidic Analysis of Phosphopeptides Using Stable Isotope-Labeled Synthetic Peptides and MRM-MS Detection,” Electrophoresis 2018. http://doi.org/10.1002/elps.201800133
- Cortes, D.F.; Tang, T.-X.; Capelluto, D.G.S.; Lazar, I.M., “Nanoflow Valve for the Removal of Trapped Air in Microfluidic Devices,” Sens. Actuators B, 2017, 243, 650–657. http://doi.org/10.1016/j.snb.2016.12.034
- Deng, J.; Lazar I.M., “Proteolytic Digestion and TiO2 Phosphopeptide Enrichment Microreactor for Fast MS Identification of Proteins,” J. Am. Soc. Mass Spectrom. 2016, 27(4), 686-698. http://doi.org/10.1007/s13361-015-1332-6
- Lazar, I.M., “Microfluidic Devices in Diagnostics: What Does the Future Hold,” Future Science 2015, 7(20), 2677-2680.
- Lazar, I.M.; Kabulski, J.L.,“Microfluidic LC Device with Orthogonal Sample Extraction for On-Chip MALDI-MS Detection,” LabChip 2013, 13(11), 2055-2065. https://doi.org/10.1039/C3LC50190F
- Armenta, J.M.; Dawoud, A.A.; Lazar, I.M., “Microfluidic Chips for Protein Differential Expression Profiling” Electrophoresis 2009, 30, 1145-1156. https://doi.org/10.1002/elps.200800653
- Dawoud, A.A.; Sarvaiya, H.A.; Lazar, I.M., “Microfluidic Platform with Mass Spectrometry Detection for the Analysis of Phosphoproteins,” Electrophoresis 2007, 28, 4645-4660. https://doi.org/10.1002/elps.200700355
- Lazar, I.M.; Trisiripisal, P.; Sarvaiya, H.A., “Microfluidic Liquid Chromatography System for Proteomic Applications and Biomarker Screening,” Anal. Chem. 2006, 78(15), 5513-5524. https://doi.org/10.1021/ac060434y
- Lazar, I.M.; Karger, B.L., “Multiple Open-Channel Electroosmotic Pumping System for Microfluidic Sample Handling,” Anal. Chem. 2002, 74(24), 6259-6268. https://doi.org/10.1021/ac0203950
- Lazar, I.M.; Sundberg, S.A.; Ramsey, R.S.; Ramsey, J.M., “Subattomole Sensitivity Microchip Nano-Electrospray Source with Time-of-Flight Mass Spectrometry Detection,” Anal. Chem. 1999, 71, 3627-3631. https://doi.org/10.1021/ac990373m
4. BIOINFORMATICS RESOURCES FOR CANCER RESEARCH
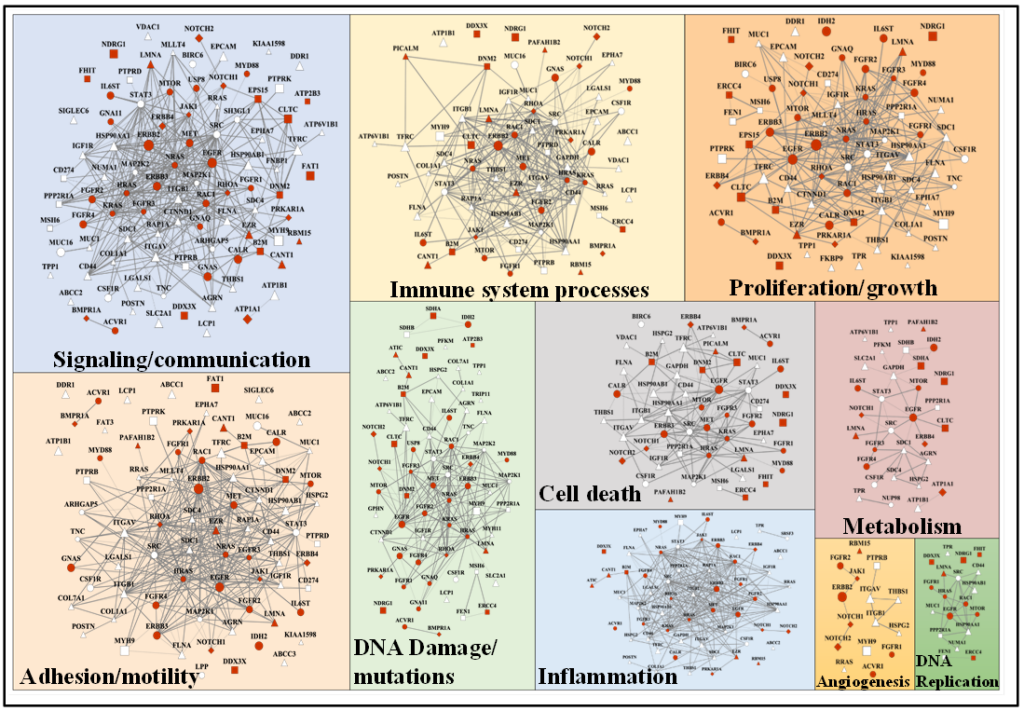
To enable cancer research, we are further interested in the development of bioinformatics resources for the analysis and interpretation of mass spectrometry data. We have developed a barcode for facilitating proteomic data normalization and differential expression analysis, a library of 20,000 peptides for biomarker and drug-target discovery, and a database encompassing over 2 million mutated peptide sequences. The top 250 mutated proteins in the database were mapped to a central cluster of intertwined biological processes that include cell development, differentiation, proliferation, response to growth factors, signaling, DNA damage sensing and repair, as well as motility and cell adhesion. As these mutated proteins are involved in some of the most significant biological processes that lead ultimately to the onset of cancer, the set represents a useful source for identifying novel networks of cancer-gene candidates, diagnostic cancer markers, and drug targets.
- Aguilera Flores, M.; Lazar, I.M., “XMAn v2 – A Database of Homo sapiens Mutated Peptides,” Bioinformatics 2020, 36(4), 1311-1313. https://doi.org/10.1093/bioinformatics/btz693
- Lazar, I.M., “Bioinformatics Resources for Interpreting Mass Spectrometry Proteomics Data,” chapter in Proteomics for Drug Discovery, book in the series of Methods in Molecular Biology, Iulia M. Lazar, Maria Kontoyianny and Alexandru C. Lazar, Eds., Springer, 2017. (ISBN 978-1-4939-7201-2; https://link.springer.com/book/10.1007/978-1-4939-7201-2
- Yang, X.; Lazar, I.M.; “XMAn: A Homo sapiens Mutated-Peptide Database for MS Analysis of Cancerous Cell States,” J. Proteome Res. 2014, 13(12), 5486-5495. https://doi.org/10.1021/pr5004467
- Lee, W.L.; Lazar, I.M., “Endogenous Protein “Barcode” for Data Validation and Normalization in Quantitative MS Analysis,” Anal. Chem. 2014, 86(13), 6379-6386. https://doi.org/10.1021/ac500855q
- Yang, X.; Lazar, I.M., “MRM Screening and Biomarker Discovery: A Library of Human Cancer-Specific Peptides” BMC Cancer 2009, 9(1), 96.https://doi.org/10.1186/1471-2407-9-96